CUT&RUN Spike-In Control Overview
CUT&RUN (Cleavage Under Targets & Release Using Nuclease) has emerged as a powerful method for investigating the genome-wide distribution of various chromatin-associated proteins and their modifications. However, the identification of differences between data sets can be challenging when global modification changes occur. Additionally inaccurate quantification of starting material or technical variation during processing results in variation across sample data. Currently available bioinformatic-based normalization methods are not applicable in these instances, and the only reliable way to overcome bias and variation is to add a known standard (Spike-In) into all samples. Active Motif offers Spike-In reagents for ChIP-Seq and CUT&Tag, and has now introduced a similar approach for CUT&RUN.
Active Motif’s strategy for CUT&RUN normalization is to Spike-In cryopreserved Drosophila cell nuclei into samples prior to CUT&RUN. Then, during the antibody incubation step, a Drosophila H2Av antibody is added in addition to the antibody to the target of interest. This Drosophila H2Av antibody provides a mechanism to reliably tag Drosophila histones in a consistent way across all samples. A normalization factor is then created based on the Drosophila signal and applied to the test genome. This CUT&RUN Spike-In strategy enables normalization of CUT&RUN data independent of the experimental antibody and without bias.
The CUT&RUN Spike-In Control works with the ChIC/CUT&RUN Assay Kit (Catalog No. 53180).
CUT&RUN Spike-In Control Highlights:
- Compare between CUT&RUN datasets between experimental samples
- Reveal differences masked by cell number inputs
- Simply add Spike-In Nuclei to samples and perform CUT&RUN with the CUT&RUN Spike-In Antibody together with experimental target antibody
CUT&RUN Spike-In Control Contents
- CUT&RUN Spike-In Antibody, store at -20°C
- CUT&RUN Spike-In Nuclei, store at -80°C
CUT&RUN Spike-In Control Data
Active Motif’s CUT&RUN normalization strategy1 may be applied to any mammalian CUT&RUN assay reaction due to the lack of cross-reactivity of the Spike-in antibody with mammalian samples. The amount of cryopreserved Drosophila nuclei and antibody used per CUT&RUN reaction may need to be optimized with the goal of having Drosophila reads make up only 5-10% of the total sequencing reads. However, when using robust antibodies against tightly localized histone modifications, such as H3K4me3, we recommend a spike-in:test sample ratio of 1:20. For antibodies against spreading marks such as H3K27me3, we recommend a spike-in:test sample ratio of 1:10. For antibodies against transcription factors such as YY1, we recommend a spike-in:test sample ratio of 1:100.
To demonstrate the utility of this approach, histone modification level differences were mimicked by setting up CUT&RUN reactions with different amounts of starting cell numbers. Various numbers of cryopreserved human K562 cells (500,000, 400,000, 300,000, 200,000, 100,000, and 50,000) were combined with 20,000 (for H3K4me3) (Figure 1) or 10,000 (for YY1) (Figure 2) of cryopreserved Drosophila nuclei for each experiment.
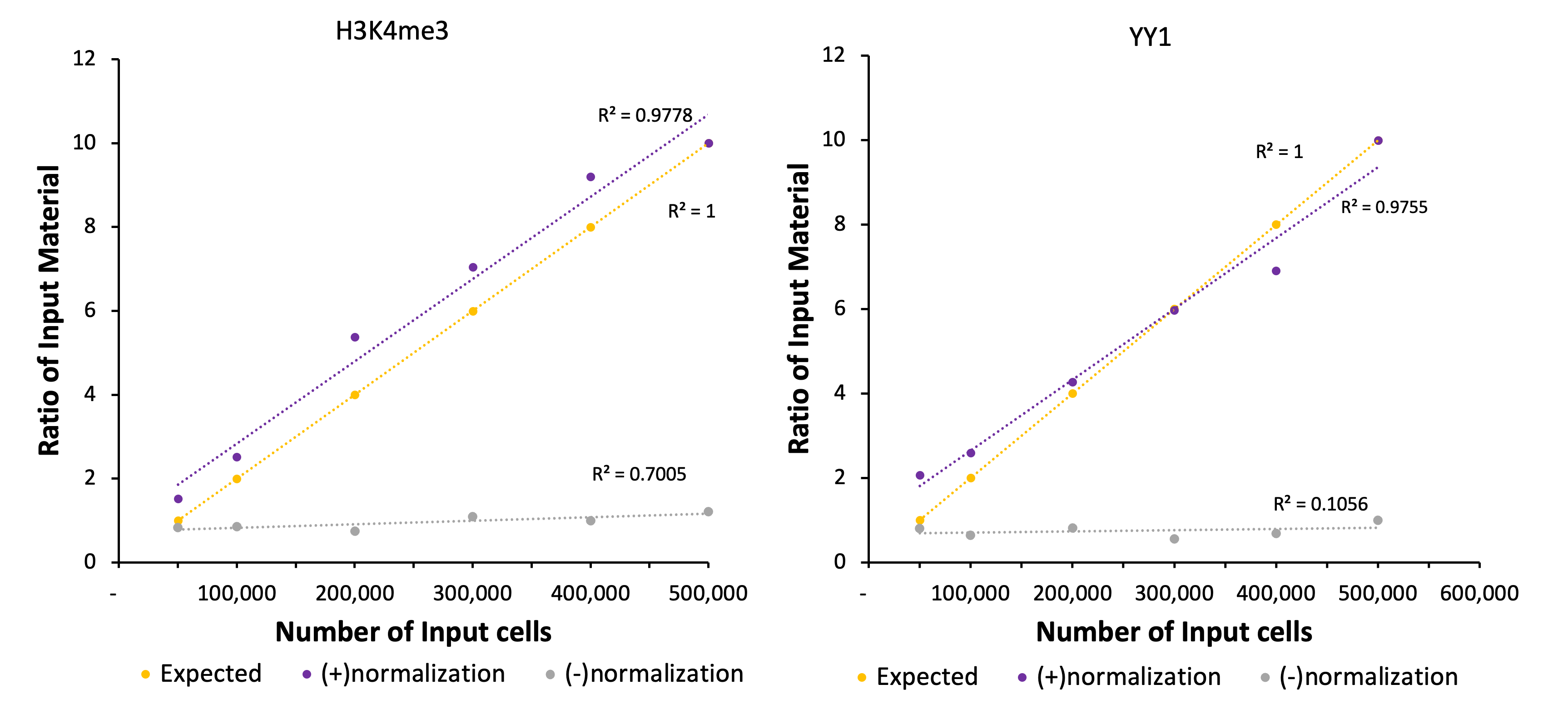
H3K4me3 and YY1, were evaluated in the CUT&RUN spike-in assay, with biological duplicates included in each experiment. Libraries were quantified and sequenced to a depth of 20-30 million reads per sample. However, sequencing to equal read depth for each sample masks the differences in starting amounts for each sample (Figures 3 and 4). Therefore spike-in normalization is required to reveal the differences in starting material2. For normalization, the sample with the lowest number of Drosophila reads was used to generate normalization factors across samples, which were then applied to down-sample the human read counts for each sample accordingly. After obtaining normalized human read counts, a standard CUT&RUN pipeline was used for peak calling generation of bigwigs. The normalized results correlate with expected results for the ratios of input material (Figures 5 and 6).
References
- Egan, B. et al. (2010) PLoS ONE. 11(11): e0166438
- Taruttis et al. (2017) Biotechniques 62:53-61
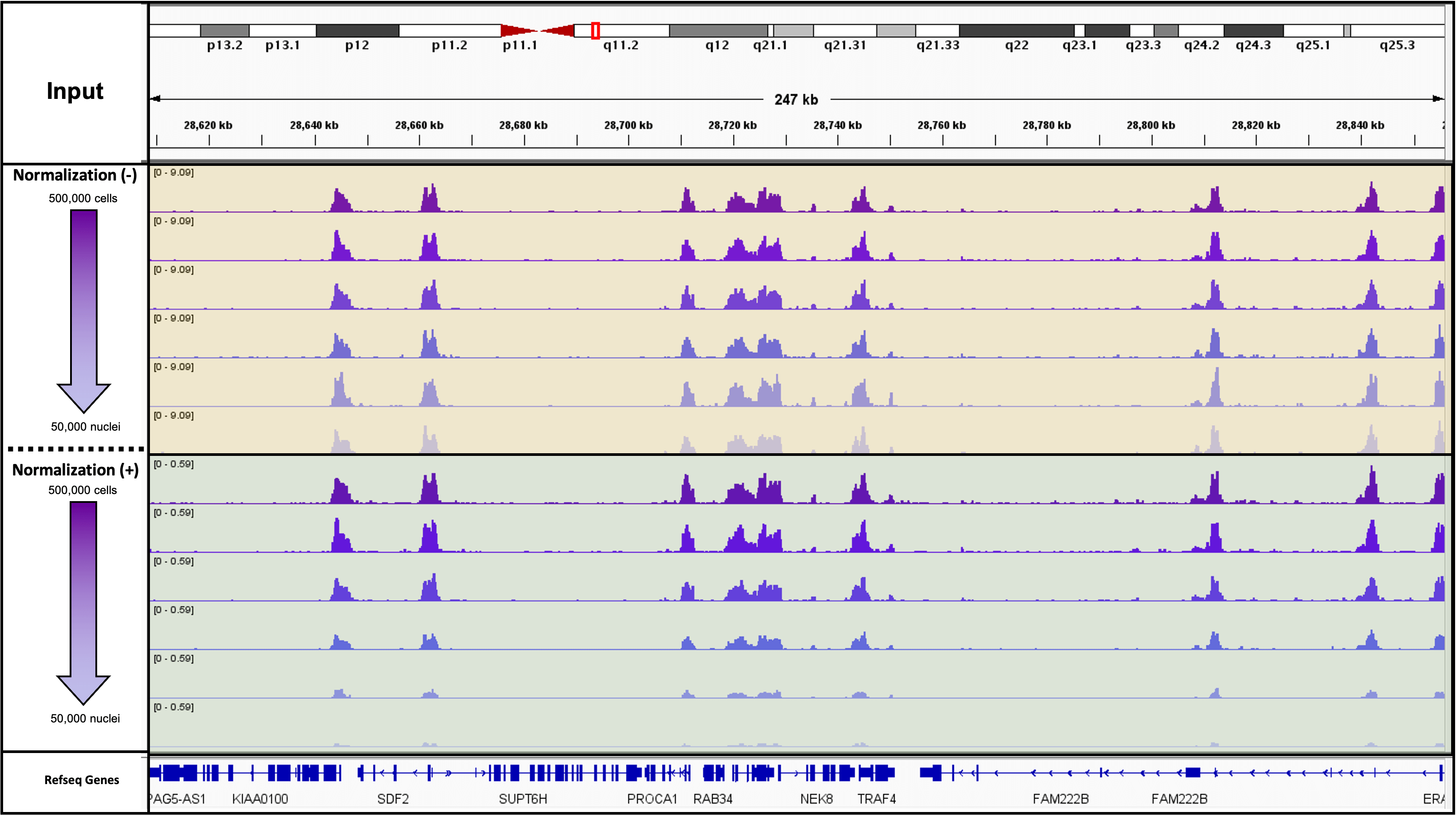
Figure 1. K562 Starting Cell Numbers for CUT&RUN Targeting H3K4me3 with and without Spike-In Normalization
20,000 CUT&RUN Spike-In Nuclei were added to 500,000, 400,000, 300,000, 200,000, 100,000 and 50,000 K562 cells and assayed in CUT&RUN. Results with and without normalization are shown.
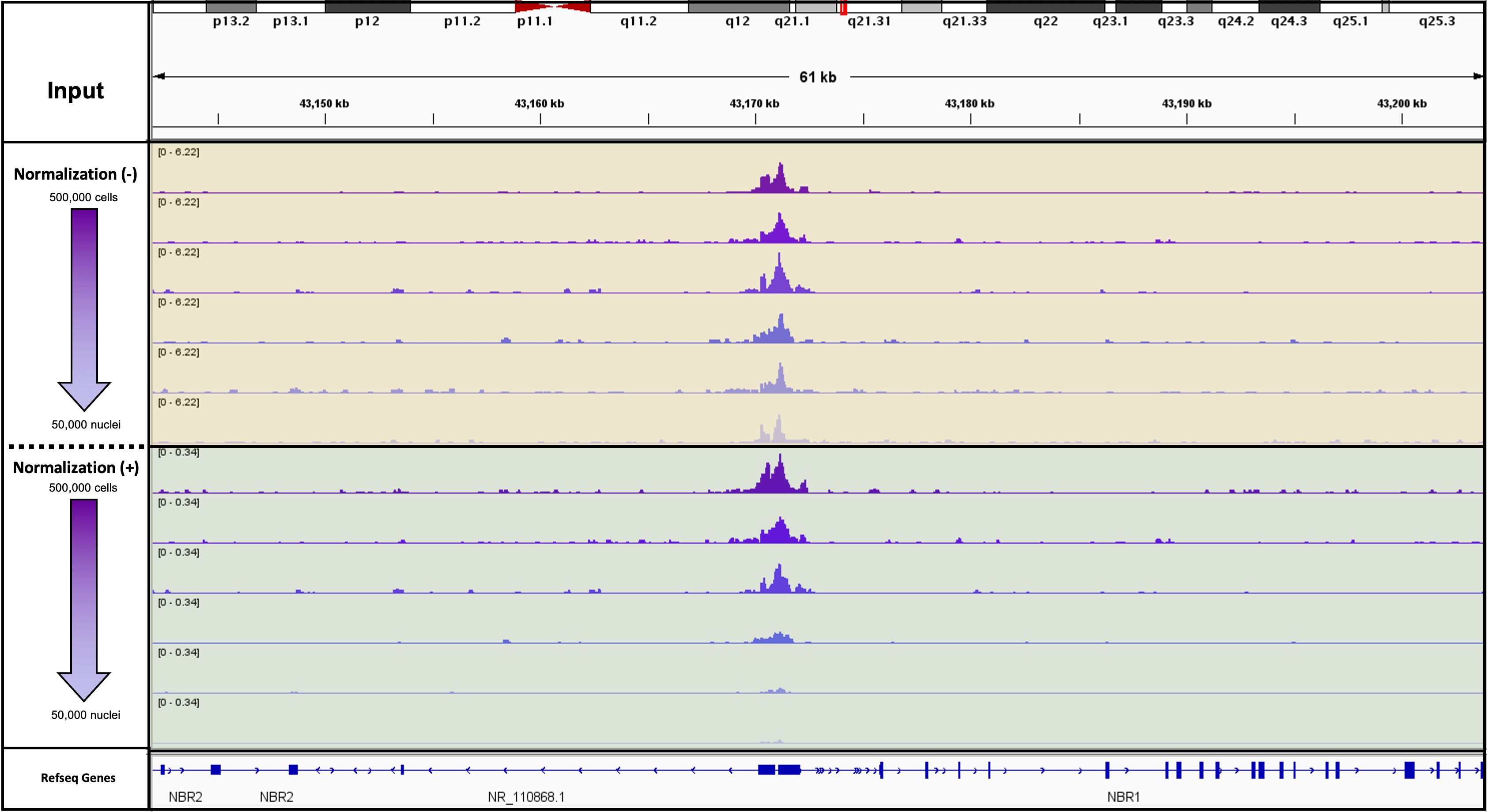
Figure 2. K562 Starting Cell Numbers for CUT&RUN targeting YY1 with and without Spike-In Normalization
5,000 CUT&RUN Spike-In Nuclei were added to 500,000, 400,000, 300,000, 200,000, 100,000 and 50,000 K562 cells and assayed in CUT&RUN. Results with and without normalization are shown.
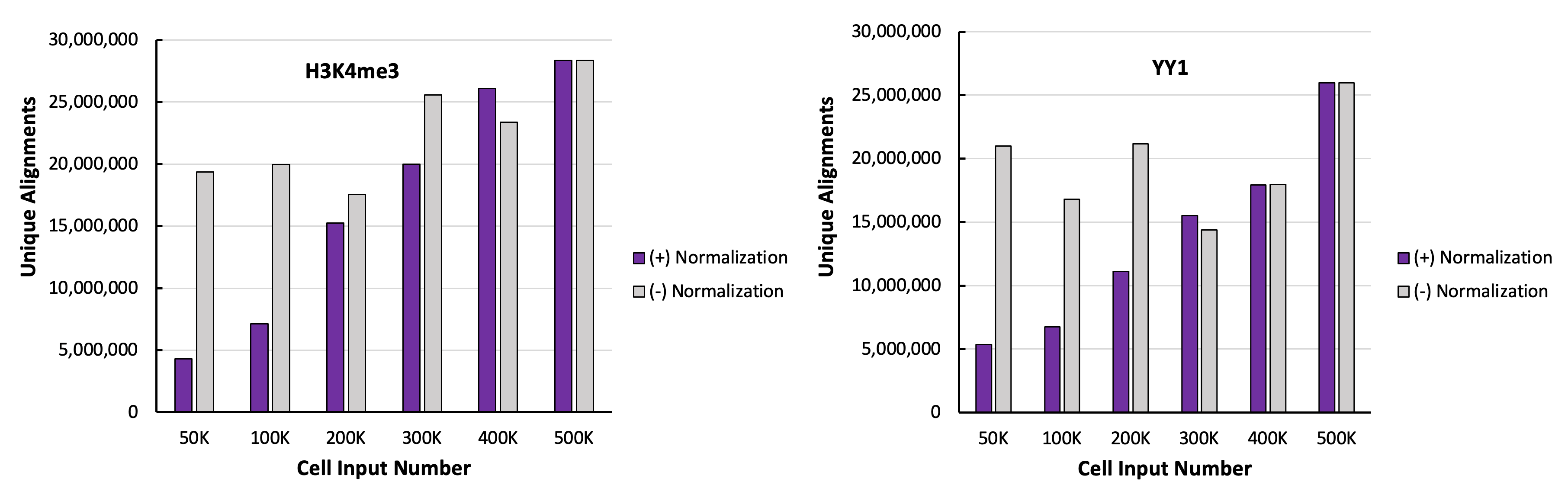
Figure 3. K562 Paired Alignments with and without Norimalization
The number of alignments is not reflective of the number of cells added to the experiments targeting H3K4me3 (left panel) and YY1 (right panel), thus normalization is needed.
Figure 4. K562 Paired Alignments After Normalization to Drosophila Paired Alignments
K562 paired alignments were normalized to the Drosophila alignment number from 500,000 K562 cells. Now the expected ratios based on amount of starting cells has been restored in H3K4me3 (left panel) and YY1 (right panel).
CUT&RUN Spike-In Control Documents
You might also be interested in:
| Name | Format | Cat No. | Price | |
|---|---|---|---|---|
| CUT&RUN Spike-In Control | 24 rxns | 53183 | ¥7,800 | Buy |