ATAC-Seq Spike-In Control Overview
Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-Seq) has emerged as a powerful method for investigating open or accessible chromatin across the genome. However, the identification of differences between data sets can be challenging when global modification changes occur, such as in the case of studying the effects of chromatin modifying enzyme inhibitors. Additionally inaccurate quantification of starting material or technical variation during processing results in variation across sample data. Currently available bioinformatic-based normalization methods are not applicable in these instances, and the only reliable way to overcome bias and variation is to add a known standard (spike-in) into all samples. Active Motif offers spike-in reagents for ChIP-Seq and CUT&Tag, and has now introduced a similar approach for ATAC-Seq.
Active Motif’s strategy for ATAC-Seq normalization is to spike-in cryopreserved Drosophila cell nuclei into samples prior to nuclei prep and tagmention. During tagmentation both the test cells and the Drosophila nuclei are tagged at open chromatin consistently across all samples. A normalization factor is then created based on the Drosophila signal and applied to the test genome.
ATAC-Seq Spike-In Control Advantages:
- Identify differences between datasets
- Reveal changes in open chromatin that were masked by cell number differences
- Overcome bias and variation with known standard
ATAC-Seq Spike-In Control Contents
- ATAC-Seq Spike-In Nuclei, store at -80°C
ATAC-Seq Spike-In Control Data
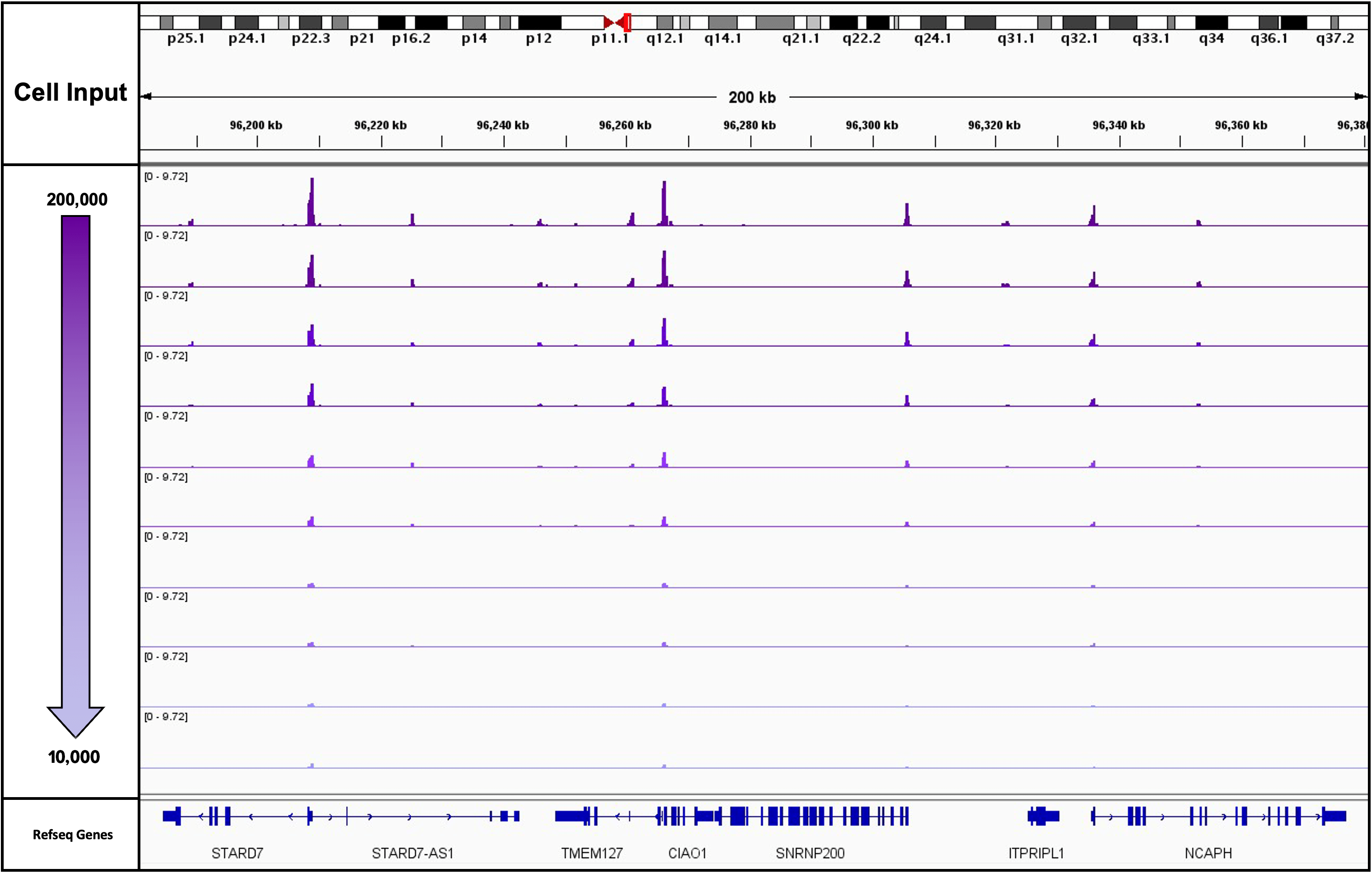
Figure 1: K562 Starting Cell Numbers for ATAC-Seq
This experiment used 10,000 drosophila nuclei as a spike in for a mock treatment. The mock treatment was a cell titration performed in duplicate from 200,000 cryopreserved K562 cells down to 10,000 cryopreserved K562 cells. The actual cell numbers are 200,000, 100,000, 50,000, 20,000 and 10,000 cells. This figure is all of the normalized tracks for the titration in duplicates.
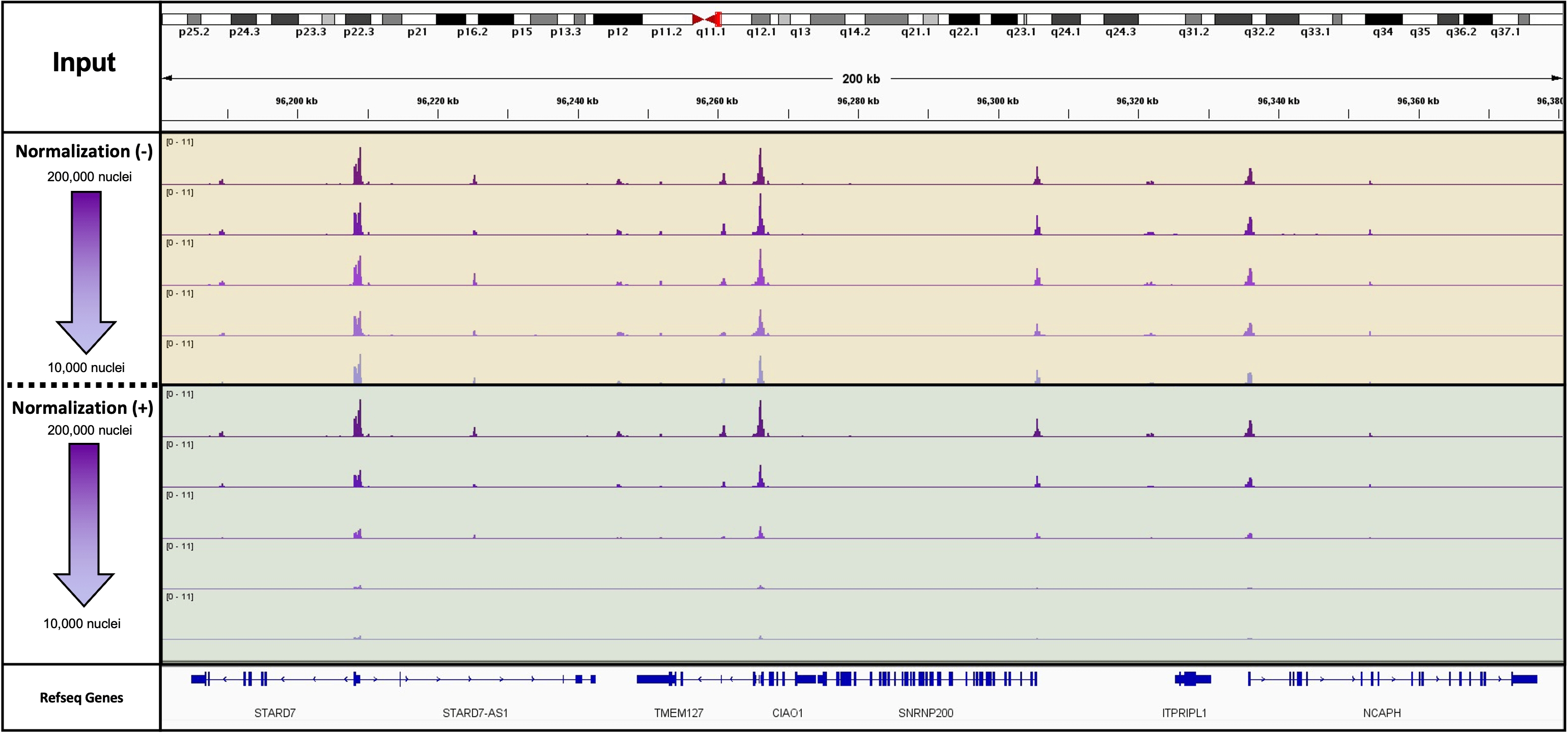
Figure 2: K562 Starting Cell Numbers for ATAC-Seq with and without Spike-In Normalization
This experiment used 10,000 drosophila nuclei as a spike in for a mock treatment. The mock treatment was a cell titration performed in duplicate from 200,000 cryopreserved K562 cells down to 10,000 cryopreserved K562 cells. The actual cell numbers are 200,000, 100,000, 50,000, 20,000 and 10,000 cells. This figure is both the spike-in normalized and unnormalized tracks for one of the duplicates in the titration.
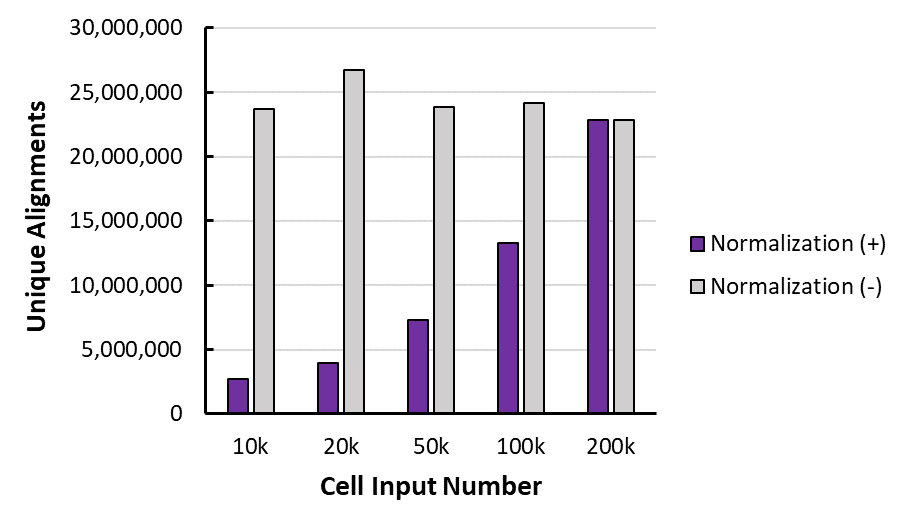
Figure 3: K562 Paired Alignments with and without Normalization
This is the unique alignments (to human genome) for one of the duplicates, before (-) and after (+) normalization with the spike in, for each of the cell inputs.
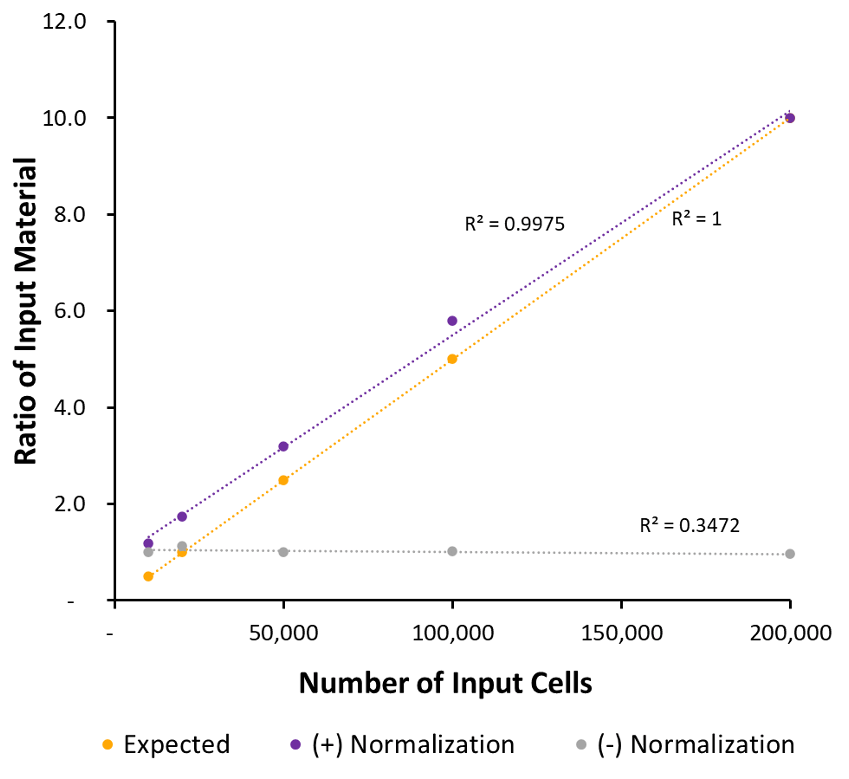
Figure 4: K562 Paired Alignments After Normalization to Drosophila Paired Alignments
This is the ratio of input material. For the sample data (normalized and unnormalized) this was calculated by dividing the number of unique fragments aligning to the human genome for each sample by the number of unique alignments for the max input. For the expected sample – the number of cell input was divided by the max number of cell input. (numbers are multiplied by 10 to increase scale).
ATAC-Seq Spike-In Control Documents
You might also be interested in:
| Name | Format | Cat No. | Price | |
|---|---|---|---|---|
| ATAC-Seq Spike-In Control | 16 rxns | 53154 | ¥5,200 | Buy |